Step 2: Matching Slices Across Stains
Aim: To identify and match structurally similar slices across stains for each patient.
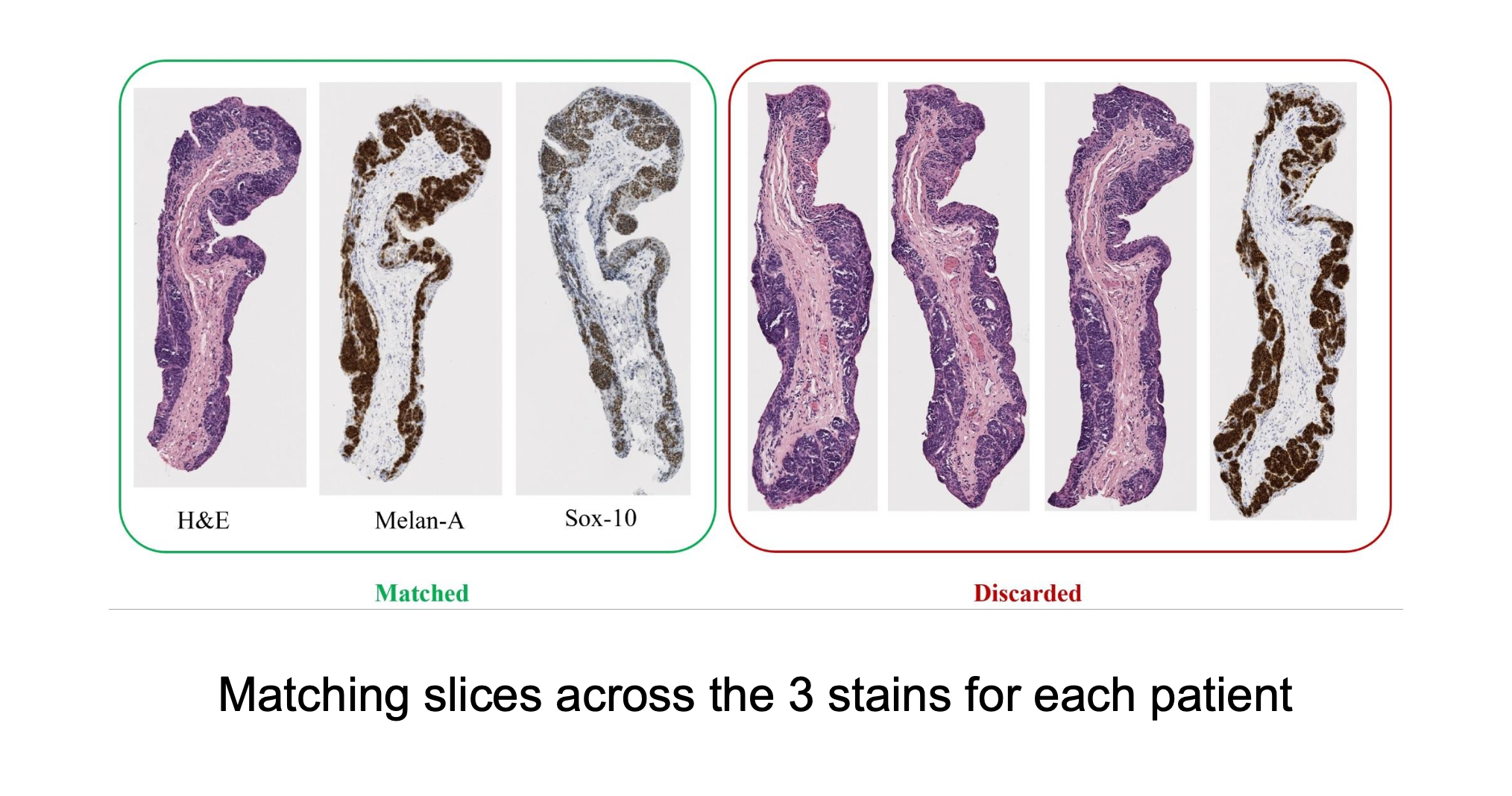
Summary
Slices from H&E, Melan-A, and SOX-10 stains are manually matched based on structural similarity and stored in labeled folders. Future automation efforts focus on contour-based matching, alignment, and patch extraction to streamline the process and ensure consistency across stains.
Methodology
The matching of slices is currently done manually. After the slices are extracted, the H&E, Melan-A, and SOX-10 slices are compared to see if there are any matches across all three stains. Often, these matches are very visually similar, but other times the silhouettes are fairly similar, which also makes for a match. Each patient’s data has its own folder, labeled by case number. Matched slices are loaded into folders named match1, match2, etc. and unmatched slices are loaded into folders named unmatched. The status of each case is seen below.
Results
Example:
All tissue slices from patient h2114153:
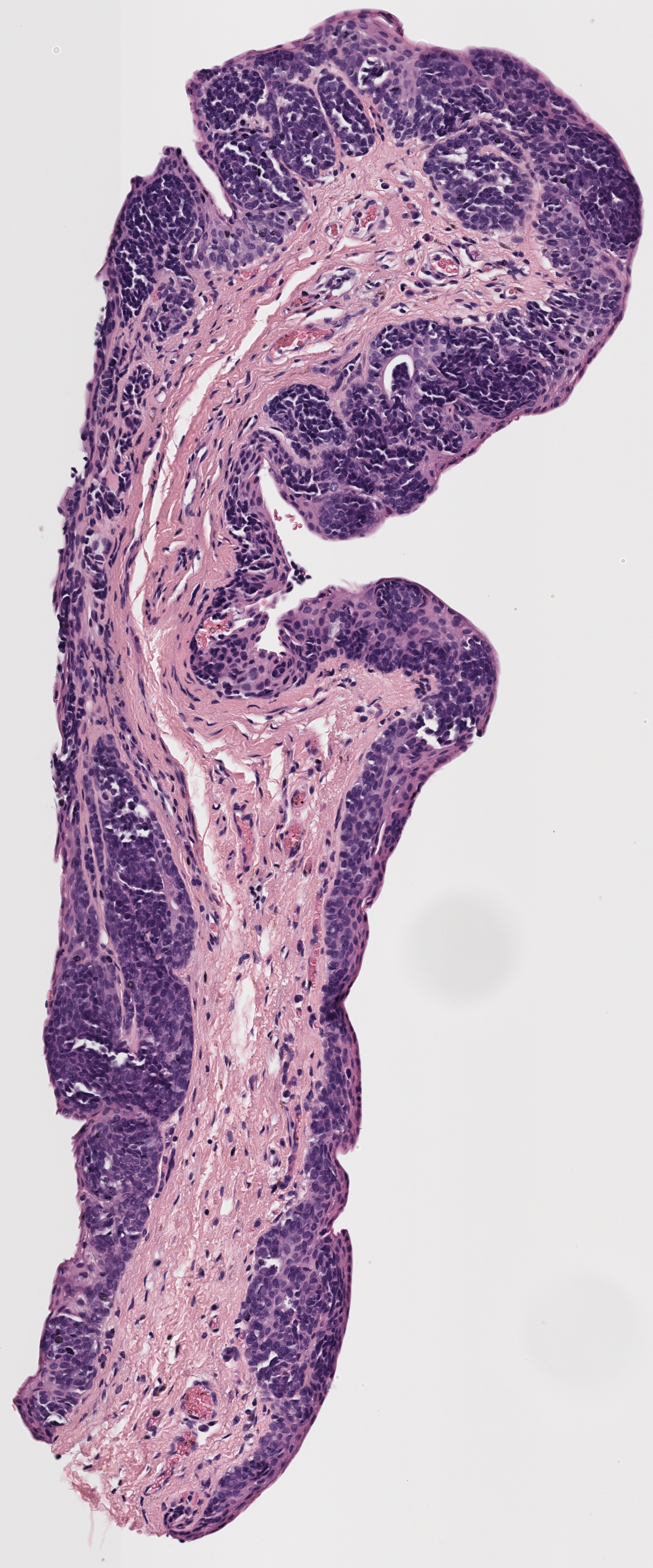
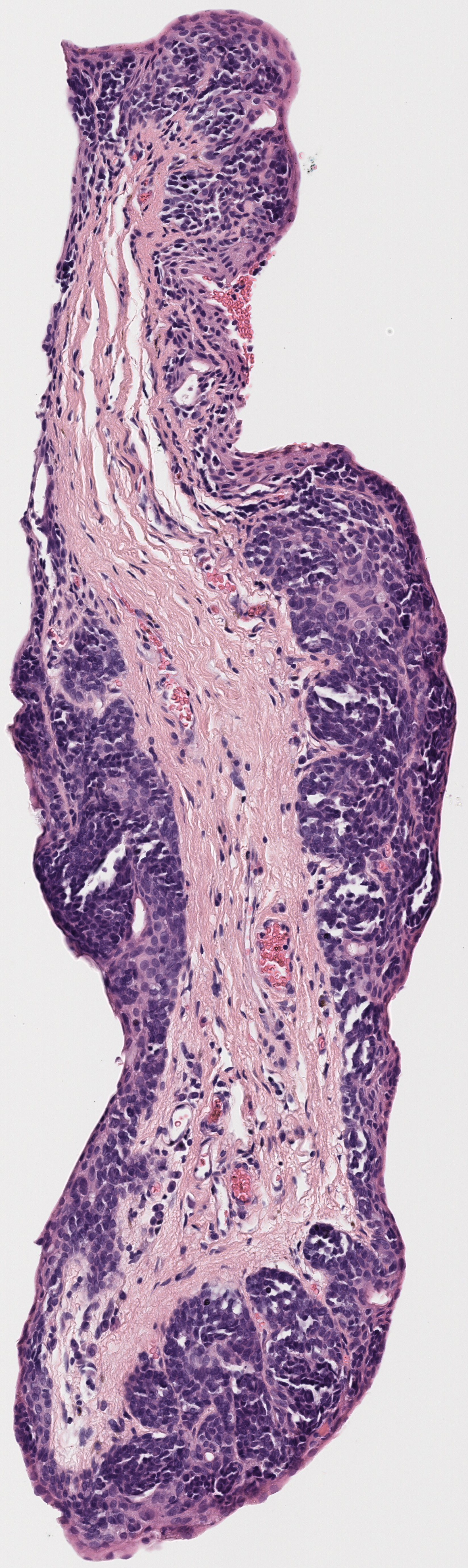
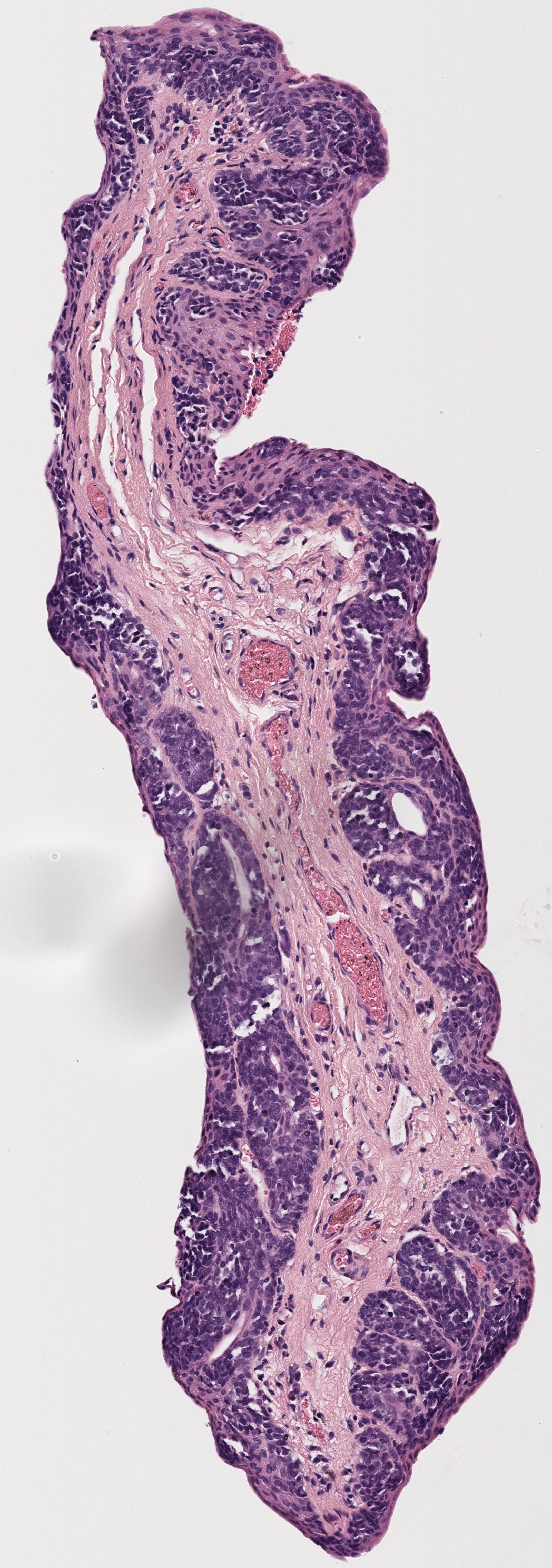
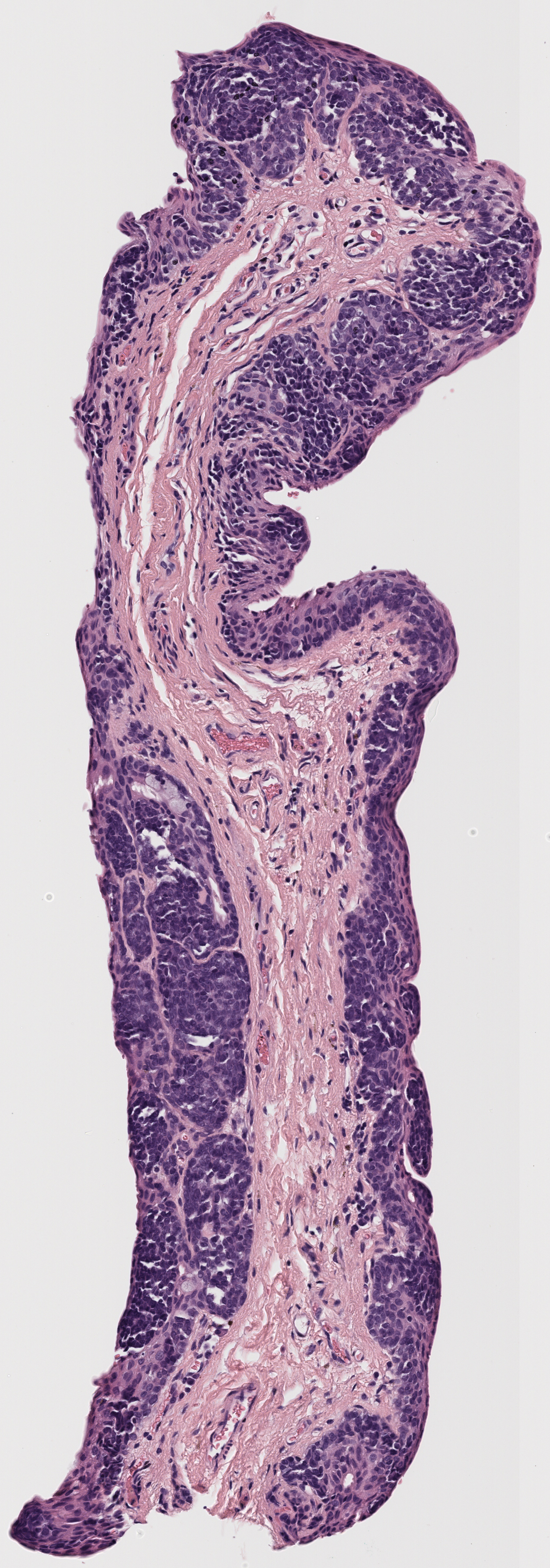
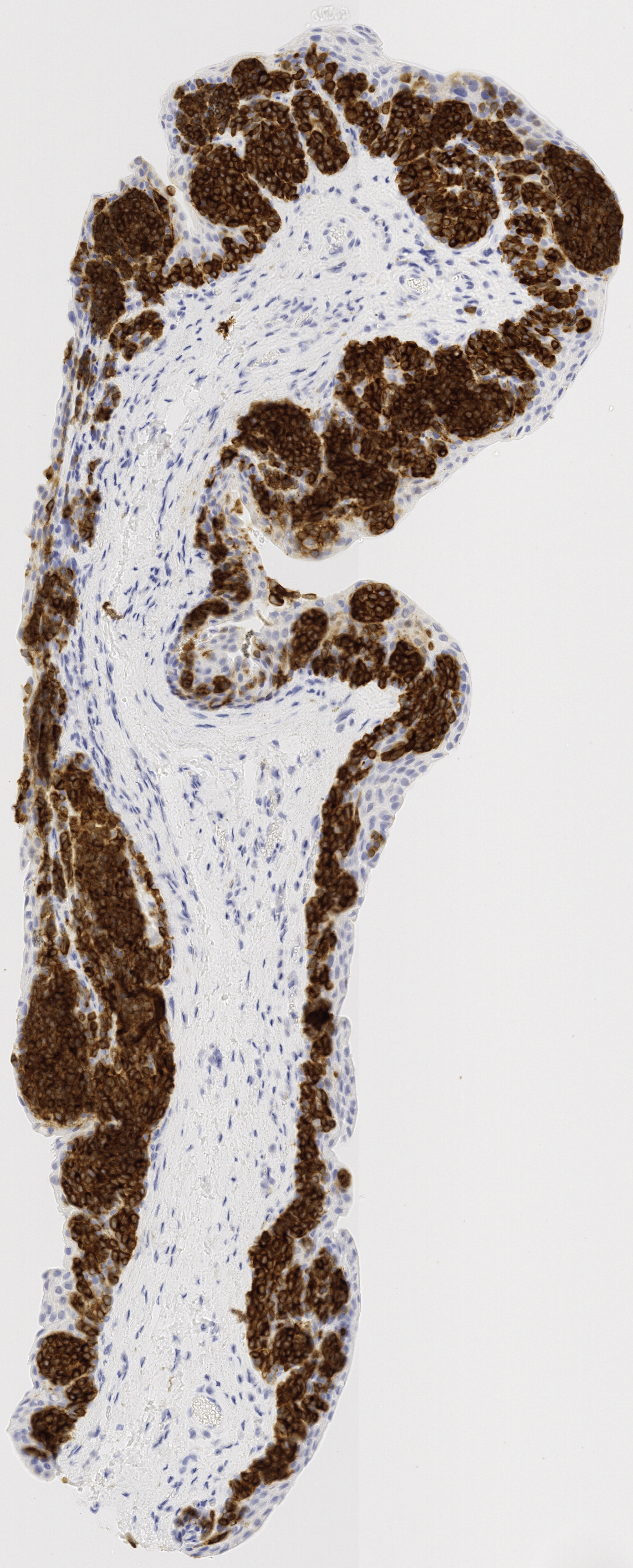
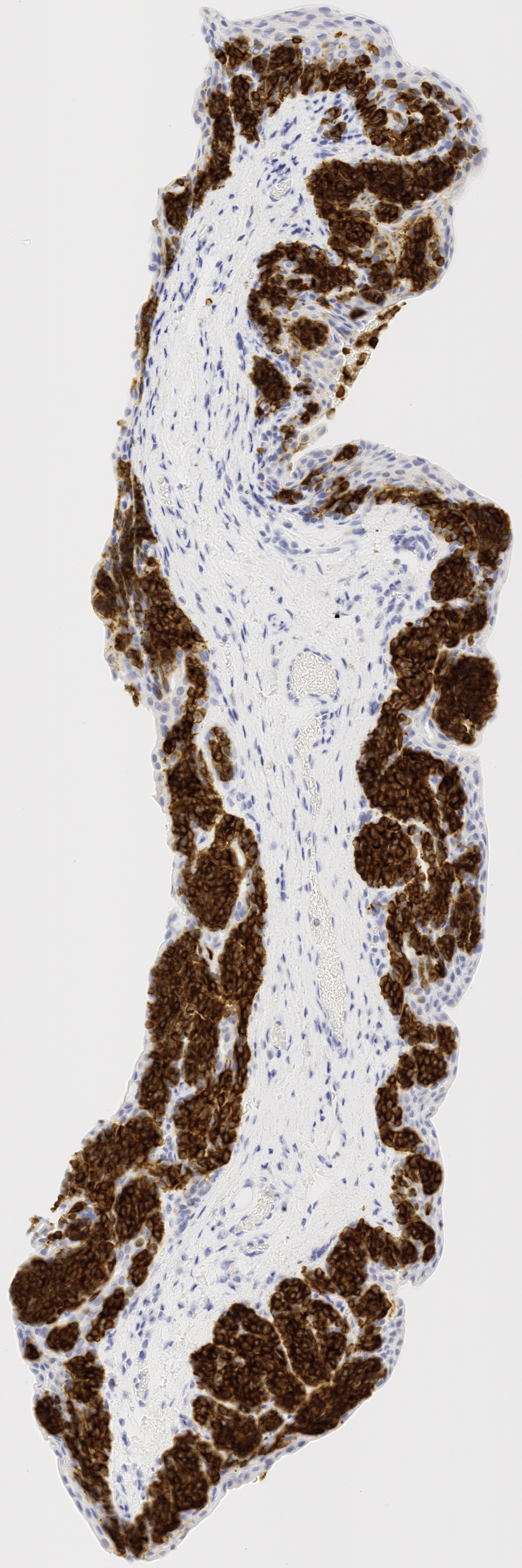
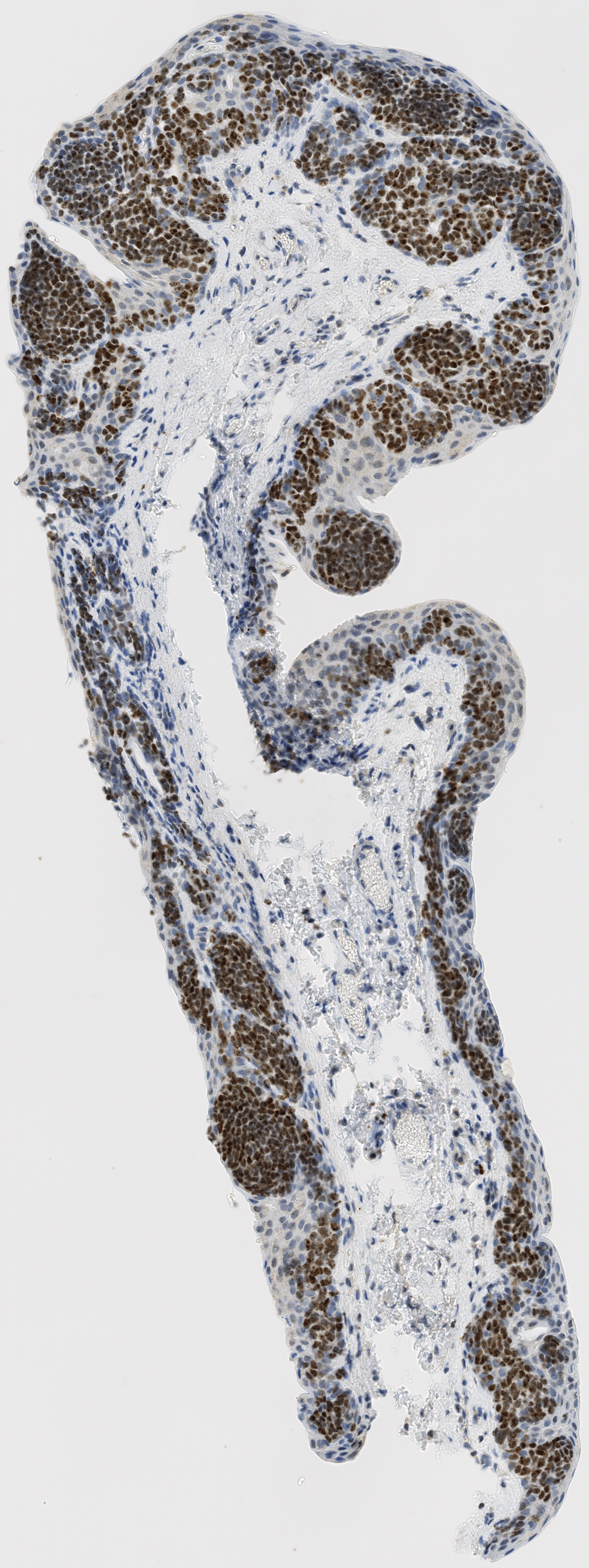
Successfully matched results




Future Automation
This pipeline automates the preprocessing, matching, alignment, and patch extraction of stained tissue images to enable efficient downstream analysis. The process begins with the preprocessing phase, where images are standardized and organized. Files are renamed to a consistent format, and patients missing one or more strain types (H&E, Melan-A, SOX-10) are excluded. For patients with multiple images of the same strain, only the highest-resolution image is retained. Once cleaned, images are split into folders by patient ID to prepare for further analysis.
The matching phase groups corresponding images across the three stains for each patient by calculating distances between extracted contours to find optimal matches. The aligned images are saved as “matches” and further processed during the alignment phase, where images are rotated and resized to maximize overlap while maintaining consistent dimensions. A contour-based algorithm crops images to their regions of interest before alignment.
Finally, the patch extraction phase identifies and extracts tissue patches containing both epithelium and stroma using skeletonization and gradient-based methods. Each patch is validated to ensure it contains components from all three stain types and meets quality criteria, such as having a balanced proportion of tissue and background pixels. Patches are saved in an organized structure, enabling seamless comparison across stains. The pipeline integrates error handling and modular design to ensure robustness, scalability, and adaptability to varied datasets.
Follow this step-by-step manual to match slices. Our API is included which automates the process.
Two codes required:
pipeline (1).py
Follow these steps to use the pipeline to generate matching slices for all patients or a specific subset:
To ensure this script works correctly, please follow the instructions below:
Run Cara’s automation script to generate the ‘processed_images’ directory
Ensure that each file is named the same way (with upper and lower case letters): patient ID + strain type + ROI number (separated by underscores)
Run the script
pipeline (1).pyand select the ‘processed_images’ directory
API
Here is the API we created to automate this step. Using this tool will speed up the process.